These pages will not reproduce all the content from the Cheetah site, but are meant as an addendum.
For a list of all Cheetah's keywords, click here.
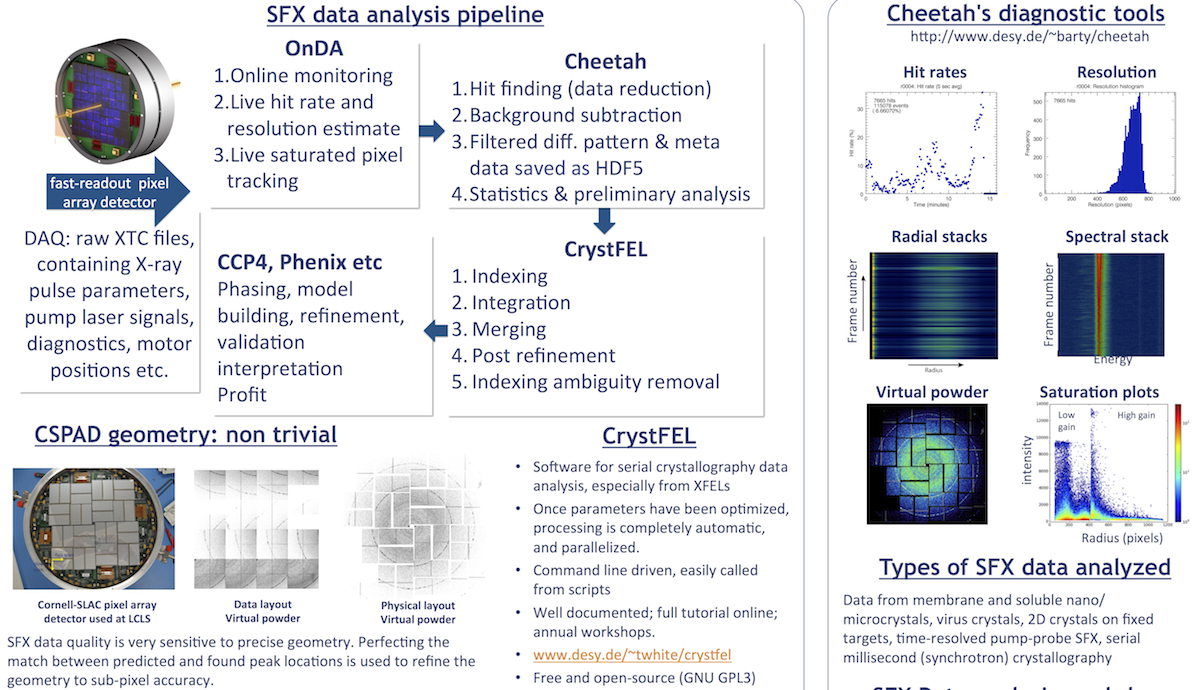
What is Cheetah?
Cheetah is a set of programs for processing serial diffraction data data from at free electron laser sources, and which enable taking home only the data with meaningful content. This is a sanity saver in many serial imaging experiments.
Cheetah is modular and can easily be adapted to any serial imaging data, including data collected using both free electron laser and synchrotron sources using a variety of detectors (including CSPAD, pnCCD, AGIPD, Pilatus, Rayonix).
The primary citation for Cheetah is:
A. Barty, R. A. Kirian, F. R. N. C. Maia, M. Hantke, C. H. Yoon, T. A. White, and H. N. Chapman, “Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data,” J Appl Crystallogr, vol. 47, pp. 1118–1131 (2014). doi:10.1107/S1600576714007626 - Download PDF - Article on IUCr website
Please cite this paper if you have used Cheetah or a part of Cheetah in your data analysis.
Downloading, compiling and installing Cheetah
Cheetah at LCLS
Step by step instructions for using the centrally installed Cheetah at LCLS: http://www.desy.de/~barty/cheetah/Cheetah/Configuration.html
Cheetah at CFEL/DESY
Cheetah is installed in /cfel/common. Running
$ cheetah-gui
should just work, provided /cfel/common/bin is in your PATH.
More instructions to follow (or ask Anton Barty)
Cheetah elsewhere
At any other location you will have to install Cheetah from scratch.Installing Cheetah itself is not too hard; however installing the LCLS framework required to read XTC files directly can be an adventure. Your mileage may vary. Please see the developer pages for details on installing Cheetah from scratch.
Alternatively, if your data comes from somewhere other than LCLS, Cheetah can be called from code able to read any other file format: it is simply a matter of passing the frame data to Cheetah for processing. Once again, see the developer pages for more details.
Cheetah for developers
Cheetah is open-source and has been released under the GNU GPL v3 license. The latest releases and updates Cheetah are best downloaded from the Github repository: https://github.com/antonbarty/cheetah/
Please follow the download instructions on that page: (assuming you have a version of git already installed)
> git clone git://github.com/antonbarty/cheetah.git
Please refer to the website for further details: http://www.desy.de/~barty/cheetah/Cheetah/Developers.html
Running Cheetah
Cheetah at LCLS
The pre-installed Cheetah package at LCLS is in /reg/g/cfel/cheetah/cheetah-latest
Please follow the instructions at http://www.desy.de/~barty/cheetah/Cheetah/Cheetah_at_LCLS.html for getting Cheetah running on your data at LCLS. Cheetah has a very handy GUI for launching batch hit finding jobs, keeping track of hit finding results, generating darkcals and bad pixel masks from them and viewing hits.
This is the most reliable route for using Cheetah at LCLS.
After you run tar -xvf /reg/g/cfel/cheetah/template.tar in your scratch/<username> directory, the sub-directories created include:
calib
Calibration files: beam (for beam files used by older versions of CrystFEL), darkcal – where you should store the darkcals created by Cheetah; gaincal – for gain calibration files; geometry – geometry files; and mask – where to store bad pixel masks, peak masks etc.
gui
Files needed by cheetah-gui. You will need to modify crawler.config before running. Instructions on what to change are on the ‘Cheetah at LCLS’ web page.
hdf5
Output from Cheetah is saved here. HDF5’s (diffraction data + metadata) and a bunch of hit finding configuration files.
A separate directory, rXXXX–<tag> is created for each run and each tag (so you can try different hit finding parameters without overwriting). The amount of “clean” data grows quickly, so remember to delete all but your best hit finding results when finished.
XXXX is the run number and <tag> is the name of your ini file if you launch jobs from the Cheetah GUI, or a user-specified tag when launching from a terminal using
./process <run> <inifile.ini> <tag>
(process, the script, can be found in your cheetah/process directory).
indexing
Location for output from CrystFEL indexing launched from the Cheetah GUI. See “lys.crystfel” script in the process directory.
process
Location for hit finding configuration (.ini) files.
- “process” sets up the environment variables and launches Cheetah
- “psana.cfg” is the configuration file for psana, the LCLS analysis framework (C++ and python).
- “lys.ini” an example .ini file.
- “darkcal.ini” – an ini file for generating a dark current measurement from a “dark” run; this doesn’t need to be edited.
The Cheetah GUI
Cheetah hit finding configuration (.ini) files
Cheetah behavior is specified by the user through a configuration file. An example configuration file, lys.ini, is provided in cheetah/process directory. Within a configuration file is a list of “keywords” that cheetah recognizes, and the user-specified values. There are two types of keywords; “global” keywords that affect the analysis of all data, and “detector” keywords that affect only one particular detector.
Global keywords may be specified in the following way:
keyword = value # comment
Note that whitespace is ignored completely, and everything following a # symbol is ignored. Keywords are not case sensitive, and if a keyword is unrecognized by Cheetah the program will exit. Look in the log file (typically .. scratch/<username>/cheetah/hdf5/rXXXX–tag/log.txt ) for which keyword was not recognized.
Detector keywords may be grouped together. One way to group keywords is to use forward slashes, as follows:
group1/keyword1 = value
group1/keyword2 = value
group2/keyword1 = value
group2/keyword2 = value
The labels group1 and group2 can be any word. An alternative way to specify groups is the following:
[group1]
keyword1 = value
keyword2 = value
[group2]
keyword1 = value
keyword2 = value
Generally, the use of brackets will simply prepend the group within the brackets to all subsequent keywords. Empty brackets are allowed, which would specify global keywords. Detector keywords that have not been assigned a group will automatically be assigned to the “first” detector.
Cheetah will ultimately be capable of performing peak finding / hit finding on multiple detectors. At the moment, these operations will only be performed on the first detector in the configuration file.
Most commonly adjusted keywords in cheetah.ini
Configure cheetah.ini by
- Selecting the right detector: see Detectors and Geometry and ask your beamline scientist to confirm.
- Selecting background processing options
- Tuning hit finding parameters
The following are the most important keywords you’ll probably ever want to tweak – the rest can likely be left alone. To read about all of Cheetah's keywords and hit finding algorithms, click here.
Detector configuration
- geometry (geometry/cspad_pixelmap.h5)
Calibration and masks
- darkcal (darkcal.h5)
- badPixelmap (badpixelmap.h5)
- peakmask (peakmask.h5)
Background subtraction
- useRadialBackgroundSubtraction (1)
- useSubtractPersistentBackground (0)
- useLocalBackgroundSubtraction (0)
Hit finding
- hitfinderADC (150)
- hitfinderMinSNR (6)
- hitfinderNPeaks (20)
- hitfinderNpeaksMax (5000)
- hitfinderMinPixCount (2)
- hitfinderMaxPixCount (20)
- hitfinderLocalBgRadius (2)
Tuning hit finding parameters
Optimising crystal hit finding
- Set hitfinderADC low enough, but not too low.
- Is there a jet streak or a bad detector region —> put it in the peak mask
- Too many spots in the solvent ring —> increase the hitfinderSNR or hitfinderMinPix
- Too few spots overall —> decrease hitfinderSNR and/or decrease the number of pixels per peak (depending on what you see for the spots not being found, too small, or too weak)
- Blank frames with little noise, finding peaks all over the place —> increase hitfinderADC (which acts as a floor on the ADC threshold computed from the radial SNR profile)
- Still stuck with too many peaks —> try restricting the radii over which hit finding is performed using hitfinderMinRes and hitfinderMaxRes (in pixels)
- It is convenient to start a new .ini file for each type of sample. The name of the .ini file is used by the GUI to tag runs and update the table, and ends up as the tag name on the HDF5 directories created. Separate names helps keep separate samples apart, and makes it easy to copy/tar/grep directories based on sample name or other eperiment parameters. This helps keep things organised. Use a symbolic link if the files are really the same.
- Review your output. Often. No analysis should ever be done completely blind. Use the ‘Show hits” button to look at images and refine the hit finding parameters.
Optimising processing speed
- Set nthreads to 16 (on LCLS and most other servers) or 72 on cfelsgi
- Check I/O speed limit using ioSpeedTest
- Turn off powder pattern creation (which skips mutex locks around summation of powder patterns)
- Increase amount of time between calculation of running background (recalculation mutex blocks all worker threads) or turn off running background completely
- Increase saveInterval
- set hitfinderFastScan to 1 – it will search only the inner 16 panels (of CSPAD’s 64)
Cheetah output files
Along with diffraction hits, virtual powder patterns and statistics, all configuration files necessary to reproduce your hitfinding result are copied into each hdf5 directory.
rXXXX-detectorX-class0-sum.h5:
Virtual powder pattern from frames not considered hits, i.e. the summation of intensities in rejected frames. Unless hdf5dump=1, frames contributing to this summation are not saved individually. This is useful to see if you are missing a lot of useful diffraction (real hits). You can view these sum.h5 files in the Cheetah GUI or using CrystFEL's hdfsee.
The HDF5 contents are explained below. Try viewing hdf5 datasets ending in “corrected_sigma” as peaks show up with much higher contrast than in the sum.
rXXXX-detectorX-class1-sum.h5:
The summation of hits, i.e. virtual powder pattern from hits.
.cxi file(s) or data1/ (data2/…) directories, containing HDF5 files
If saveCXI=1 (default), all hits and corresponding metadata are saved in CXI format, i.e. in a single, large HDF5 as described in https://github.com/FilipeMaia/CXI/raw/master/cxi_file_format.pdf
If you set saveCXI=0 in the .ini file, individual HDF5’s are saved in data directories of up to 1000 small HDF5 files each. HDF5 filename: LCLS_year_monthday_rXXXX_hhmmss_tttt.h5
A short description of the HDF5 file content / structure can be found further below.
If you ran darkcal.ini, no data1 etc directories will be created. The dark current measurement (averaged over the whole dark run) will be in a file called cxiXXXXX-rXXXX-detectorX-darkcal.h5. Copy this to your cheetah/calib/darkcal directory (feel free to rename it, but keep track of which run it was from and which detector, if using multiple detectors), for easier reference. Update your ini files to point to the new darkcal. Always use the dark cal nearest to your sample runs. If in doubt, use a later one.
A cxiXXXX-rXXXX.cxi file with shot-by-shot metadata will still be created even if you run a darkcal.ini. You have view its contents in hdfsee or some other HDF5 viewer, or h5dump -d <dataset name>. (Run h5dump -n <file.cxi> first to see what the datasets are, before trying to dump potentially GB's of text).
frames.txt:
Frames.txt contains a list of all detector readout events (hits and non-hits) with various attributes.
eventData->________ meaning:
- eventName HDF5 filename: LCLS_year_monthday_rXXXX_hhmmss_tttt.h5 (if saveCXI=0)
- filename “---“ if non hit; “data*/filename “ if hit.
- stackSlice
- xtcFrameNumber
- hit
- powderClass 0 = non hits; 1 = hits
- hitScore
- photonEnergyeV
- wavelength Å
- gmd1 gas monitor detector 1 (for incident flux measurement)
- gmd2 gas monitor detector 2 (for incident flux measurement)
- detector[0].detector sum of “detectorZpvname” and “cameraLengthOffset” in .ini file
- energySpectrumExist was the spectrometer in place and recorded to datastream?
- nPeaks Number of peaks found
- peakNpix total number of pixels that contribute to peaks in pattern
- peakTotal total intensity of all peak pixels
- peakResolution in pixels
- peakDensity
- pumpLaserCode process variable where laser trigger is recorded (evr41, evr183, LD57)
- pumpLaserDelay
- pumpLaserOn trigger for pump laser experiments
cleaned.txt
Cleaned.txt contains information about only the hits, with fewer columns than frames
- Filename info->eventname
- frameNumber threadNum
- npeaks info->nPeaks
- nPixels info->peakNpix
- totalIntensity info->peakTotal
- peakResolution info->peakResolution (pixels)
- peakResolutionA info->peakResolutionA
- peakDensity info->peakDensity
rXXXX-class0-log.txt and rXXXX-class1-log.txt
Lists of files contibuting to rXXXX-detectorX-classX-sum.h5.
Similar to frames.txt and cleaned.txt but class0 = non hits, and class1 = hits. (so cleaned.txt and class1-log.txt will contain the same files). The columns are:
eventData->eventname, eventData->filename, eventData->stackSlice, eventData->xtcFrameNumber, eventData->hitScore, eventData->photonEnergyeV, eventData->wavelengthA, eventData->detector[0].detectorZ, eventData->gmd1, eventData->gmd2, eventData->energySpectrumExist, eventData->nPeaks, eventData->peakNpix, eventData->peakTotal, eventData->peakResolution, eventData->peakDensity, eventData->pumpLaserCode, eventData->pumpLaserDelay
darkcal.h5
Copy of the dark current measurement specified in original.ini as darkcal.
geometry.h5
Copy of pixel map specified in the .ini file under geometry.
Log.txt
Progress of hit finding, updated at the rate set by keyword saveInterval. If hit finding has finished, a summary is appended, including total frames processed, number of hits, hit rate, average photon energy and its sigma.
bsub.log
Log from batch job submission. Look here for errors when hit finding doesn’t work. It will report misuse of keywords and other problems.
original.ini
Copy of your original ini file (renamed).
cheetah.ini
Same as original.ini but with commented lines removed.
cheetah.out
Full list of parameters used by Cheetah for this hit finding. You can see here if any of your keyword values from cheetah.ini were overwritten automatically due to clashes.
Peakmask.h5
Copy of mask used while peak finding. See keyword peakmask.
Peaks.txt
Space separated column file with peak information. One line per peak.
frameNumber, eventName, photonEnergyEv, wavelengthA, GMD, peak_index, peak_x_raw, peak_y_raw, peak_r_assembled, peak_q, peak_resA, nPixels, totalIntensity, maxIntensity, sigmaBG, SNR
Where GMD is a gas monitoring detector (proportional to incident flux), and peak_index: is
Psana.cfg
Copy of psana.cfg from your cheetah/process directory: the configuration file for psana, the LCLS analysis framework.
Status.txt
Status of hit finding.
xtcfiles.txt
List of xtc files from this run. If you started your hit finding before all the xtc files finished writing to the offline storage, this list may be incomplete. You will not see an error in the output from Cheetah if everything runs correctly, but rerunning it at a later date will show more frames processed. In the directory where the raw data are saved, /reg/d/psdm/cxi/cxiXXXXX/xtc (XXXXX = the experimental ID with the last 2 digits corresponding to the year of the experiment), while the xtc files are being written, their names are appended with .inprogress and Cheetah deliberately excludes them until the extension is solely .xtc.
Cheetah output HDF5 contents
This is what may be referred to as "cleaned" data.
In serial femtosecond crystallography, you will typically hit each crystal only once. (Unless the crystals are large and flowing slowly). Each diffraction pattern that Cheetah finds in the raw data stream from LCLS is saved as an individual HDF5 if the keyword "saveCXI = 0". If saveCXI =1, all the hits are saved into one large HDF5 file in "CXI format" as described in https://github.com/FilipeMaia/CXI/raw/master/cxi_file_format.pdf
HDF5 files are hierarchical, consisting of the groups and dataset. Groups can contain other groups and datasets, while datasets can contain multi-dimensional data (e.g. diffraction data). More on HDF5's on the HDF5 page.
If saveCXI=0
Each HDF5 file will have the following contents (called datasets. links are allowed within an HDF5 too...)
|
LCLS_2013_Feb12_r0194_053144_17343.h5
|
|
/LCLS
|
/data
|
/processing
|
|
|
/detector0-EncoderValue
|
|
/data
|
|
/energySpectrum-tilt
|
|
|
/detector0-Position
|
|
/energySpectrum1D
|
|
/hitfinder
|
|
|
/detector1-EncoderValue
|
|
/energySpectrumCCD
|
|
|
/peakinfo
|
|
|
/detector1-Position
|
|
/energySpectrumScale
|
|
|
/peakinfo-assembled
|
|
|
/ebeamCharge
|
|
/radialAverage0
|
|
|
/peakinfo-raw
|
|
|
/ebeamL3Energy
|
|
/radialAverage1
|
|
/pixelmasks
|
|
|
/ebeamLTUAngX
|
|
/radialAverageCounter0
|
|
|
|
|
/ebeamLTUAngY
|
|
/radialAverageCounter1
|
|
|
|
|
/ebeamLTUPosX
|
|
/rawdata
|
|
|
|
|
/ebeamLTUPosY
|
|
/rawdata0
|
|
|
|
|
/ebeamPkCurrBC2
|
|
/rawdata1
|
|
|
|
|
/eventTimeString
|
|
|
|
|
|
|
/evr41
|
|
|
|
|
|
|
/f_11_ENRC
|
|
|
|
|
|
|
/f_12_ENRC
|
|
|
|
|
|
|
/f_21_ENRC
|
|
|
|
|
|
|
/f_22_ENRC
|
|
|
|
|
|
|
/fiducial
|
|
|
|
|
|
|
/machineTime
|
|
|
|
|
|
|
/phaseCavityCharge1
|
|
|
|
|
|
|
/phaseCavityCharge2
|
|
|
|
|
|
|
/phaseCavityTime1
|
|
|
|
|
|
|
/phaseCavityTime2
|
|
|
|
|
|
|
/photon_energy_eV
|
|
|
|
|
|
|
/photon_wavelength_A
|
|
|
|
|
This table shows the structure of the virtual powder patterns, rXXXX-detectorX-classX-sum.h5
|
R0016-detector0-class1-sum.h5
|
|
/data (group)
|
/data (link)
|
|
|
/nframes
|
|
/correcteddata --> /data/non_assembled_detector_corrected
|
|
|
/non_assembled_detector_and_photon_corrected
|
|
/data --> /data/non_assembled_detector_corrected
|
|
|
/non_assembled_detector_and_photon_corrected_sigma
|
|
|
|
|
/non_assembled_detector_corrected
|
|
|
|
|
/non_assembled_detector_corrected_sigma
|
|
|
|
|
/peakpowder
|
|
|
|
|
/radial_average_detector_and_photon_corrected
|
|
|
|
|
/radial_average_detector_and_photon_corrected_sigma
|
|
|
|
|
/radial_average_detector_corrected
|
|
|
|
|
/radial_average_detector_corrected_sigma
|
|
|
Other 2D datasets may be created when other "savePowder" keywords are set to 1 in the ini file.
If saveCXI=1 (default)
All hits and corresponding metadata are saved in CXI format in a single HDF5. The structure of this .cxi file is described in CXI stack of images from a modular detector in the CXI format documents in www.cxidb.org.
Cheetah GUI
For LCLS users:
The Cheetah website has detained instructions for setting up your experimental directories to use the centrally installed Cheetah: http://www.desy.de/~barty/cheetah/Cheetah/Configuration.html
Scripts
Scripts to help with miscellaneous tasks while hit finding and doing preliminary analysis can be downloaded from https://www.bioxfel.org/resources/scripts
peakogram
Quickly plot a histogram of all peaks found in run. Good way to see if you have a lot of saturation and to estimate resolution from the whole run. Uses peaks.txt.
A script to quickly calculate hits and hitrates from SFX experiments
A script to quickly calculate data quality metrics from SFX experiments
visibly_bad_mask.py
A script to generate masks (bad pixel or peak) manually. Useful for shadows, rings from substrate/sample holder. The resultant binary (0/1) hdf5 mask needs<