Popular Articles
- Earliest molecular events of vision revealed
- Dynamics and Kinetics in Structural Biology
- XFEL Pulses Demonstrate How Plants Perceive Light
- Structural biology is solved -- now what?
- BioXFEL Postdoctoral Fellowship Award
Archived Articles
- Details
- Thursday, 04 November 2021

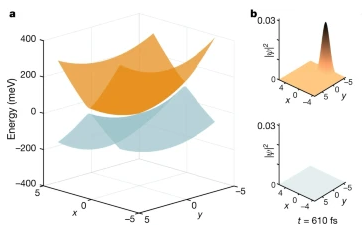
The structural dynamics of a molecule are determined by the underlying potential energy landscape. Conical intersections are funnels connecting otherwise separate potential energy surfaces. Posited almost a century ago, conical intersections remain the subject of intense scientific interest.
In biology, they have a pivotal role in vision, photosynthesis and DNA stability6. Accurate theoretical methods for examining conical intersections are at present limited to small molecules. Experimental investigations are challenged by the required time resolution and sensitivity. Current structure-dynamical understanding of conical intersections is thus limited to simple molecules with around ten atoms, on timescales of about 100 fs or longer. Spectroscopy can achieve better time resolutions, but provides indirect structural information.
Here we present few-femtosecond, atomic-resolution videos of photoactive yellow protein, a 2,000-atom protein, passing through a conical intersection. These videos, extracted from experimental data by machine learning, reveal the dynamical trajectories of de-excitation via a conical intersection, yield the key parameters of the conical intersection controlling the de-excitation process and elucidate the topography of the electronic potential energy surfaces involved.
View the full article:
Hosseinizadeh, A., Breckwoldt, N., Fung, R. et al. Few-fs resolution of a photoactive protein traversing a conical intersection. Nature (2021).
https://doi.org/10.1038/s41586-021-04050-9





